New Caledonia¶
Downloading data in parallel¶
We can use geefcc to download forest cover change for large countries, for example New-Caledonia. The country will be divided into several tiles which are processed in parallel. If your computer has n cores, n-1 cores will be used in parallel.
import os
import time
import ee
import numpy as np
import pandas as pd
from osgeo import gdal
from geefcc import get_fcc, sum_raster_bands
import geopandas
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
import matplotlib.patches as mpatches
from tabulate import tabulate
We initialize Google Earth Engine.
# Initialize GEE
ee.Initialize(project="deforisk",
opt_url=("https://earthengine-highvolume."
"googleapis.com"))
We can compute the number of cores used for the computation.
ncpu = os.cpu_count() - 1
ncpu
7
Using TMF product¶
Downloading data¶
We download the forest cover change data from GEE for New Caledonia for years 2001, 2010 and 2020, using a tile size of one degree. We use the TMF product (version v1_2023 available in geefcc).
start_time = time.time()
get_fcc(
aoi=(163.5, -23, 168.15, -19.51),
buff=0.0,
years=[2001, 2010, 2020],
source="tmf",
tile_size=1.0,
crop_to_aoi=True,
output_file="out_tmf/forest_tmf.tif",
)
end_time = time.time()
We estimate the computation time to download 20 1-degree tiles using several cores.
elapsed_time = (end_time - start_time) / 60
print('Execution time:', round(elapsed_time, 2), 'minutes')
Execution time: 9.55 minutes
Transform multiband fcc raster in one band raster¶
We transform the data to have only one band describing the forest cover change with 0 for non-forest, 1 for deforestation on the period 2000–2009, 2 for deforestation on the period 2010–2019, and 3 for the remaining forest in 2020. To do so, we just sum the values of the raster bands.
sum_raster_bands(input_file="out_tmf/forest_tmf.tif",
output_file="out_tmf/fcc_tmf.tif",
verbose=False)
We resample at a lower resolution for plotting.
infn = "out_tmf/fcc_tmf.tif"
outfn = "out_tmf/fcc_tmf_coarsen.tif"
scale = gdal.Open(infn).GetGeoTransform()[1]
xres = 20 * scale
yres = 20 * scale
resample_alg = "near"
ds = gdal.Warp(outfn, infn, xRes=xres, yRes=yres, resampleAlg=resample_alg)
ds = None
Plot the forest cover change map¶
We prepare the colors for the map.
# Colors
cols=[(255, 165, 0, 255), (227, 26, 28, 255), (34, 139, 34, 255)]
colors = [(1, 1, 1, 0)] # transparent white for 0
cmax = 255.0 # float for division
for col in cols:
col_class = tuple([i / cmax for i in col])
colors.append(col_class)
color_map = ListedColormap(colors)
# Labels
labels = {0: "non-forest in 2001", 1:"deforestation 2001-2009",
2:"deforestation 2010-2019", 3:"forest in 2020"}
patches = [mpatches.Patch(facecolor=col, edgecolor="black",
label=labels[i]) for (i, col) in enumerate(colors)]
We load the data: country borders and grid. The borders can be downloaded from the gadm website.
# Borders
borders_gpkg = os.path.join("data", "borders_NCL.gpkg")
borders = geopandas.read_file(borders_gpkg)
# Grid
grid_gpkg = os.path.join("out_tmf", "grid.gpkg")
grid = geopandas.read_file(grid_gpkg)
We plot the forest cover change map.
with gdal.Open("out_tmf/fcc_tmf_coarsen.tif", gdal.GA_ReadOnly) as ds:
raster_image = ds.ReadAsArray()
nrow, ncol = raster_image.shape
xmin, xres, _, ymax, _, yres = ds.GetGeoTransform()
extent = [xmin, xmin + xres * ncol, ymax + yres * nrow, ymax]
# Plot
fig = plt.figure()
ax = plt.subplot(111)
ax.imshow(raster_image, cmap=color_map, extent=extent,
resample=False)
ax.set_aspect("equal")
grid_image = grid.boundary.plot(ax=ax, color="grey", linewidth=0.5)
borders_image = borders.boundary.plot(ax=ax, color="black", linewidth=0.5)
plt.title("Forest cover change 2001-2010-2020, TMF")
plt.legend(handles=patches, bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.xlim((163, 169))
fig.savefig("fcc_tmf.png", bbox_inches="tight", dpi=200)
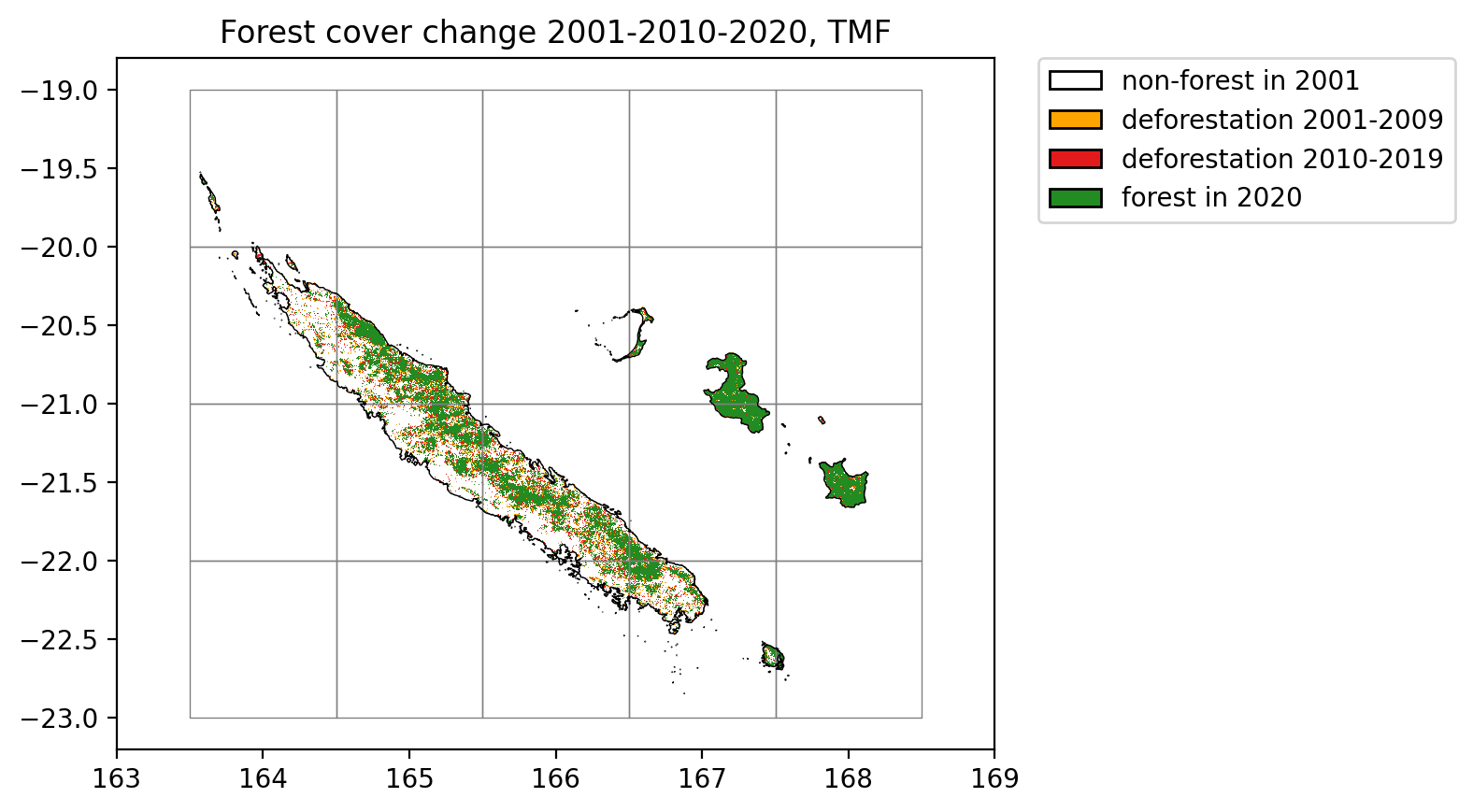
Lines in black represent country borders and the 10 km buffer. One degree tiles in grey cover the whole buffer and were used to download the data in parallel.
Reproject in EPSG:3163 for area computation¶
ifile = os.path.join("out_tmf", "fcc_tmf.tif")
ofile = os.path.join("out_tmf", "fcc_tmf_epsg3163.tif")
ds = gdal.Warp(ofile, ifile, xRes=30, yRes=30, dstSRS="EPSG:3163", resampleAlg="near",
targetAlignedPixels=True, creationOptions=["COMPRESS=DEFLATE"])
ds = None
Compute statistics¶
We use the tool “Raster layer unique values report” in QGIS to get the number of pixels per pixel value in the raster.
pixel_count = [n1, n2, n3] = [273463, 257445, 9402021]
areas = [round(i * (30 * 30 / 10000)) for i in pixel_count]
tmf_areas= {"product": "tmf", "version": "v1_2023", "perc": "",
"fc2001": areas[0] + areas[1] + areas[2],
"fc2010": areas[1] + areas[2], "fc2020": areas[2],
"d1": round(areas[0] / 9), "d2": round(areas[1] / 10)}
print(tmf_areas)
{'product': 'tmf', 'version': 'v1_2023', 'perc': '', 'fc2001': 893964, 'fc2010': 869352, 'fc2020': 846182, 'd1': 2735, 'd2': 2317}
Using GFC product and tree cover \(\ge\) 80%¶
Downloading data¶
We download the forest cover change data from GEE for New Caledonia for years 2001, 2010 and 2020, using a tile size of one degree. We use the GFC product (version v1_11 available in geefcc) and a tree cover percentage \(\ge\) 80 to define the forest.
start_time = time.time()
get_fcc(
aoi=(163.5, -23, 168.15, -19.51),
buff=0.0,
years=[2001, 2010, 2020],
source="gfc",
perc=80,
tile_size=1.0,
crop_to_aoi=True,
output_file="out_gfc80/forest_gfc80.tif",
)
end_time = time.time()
We estimate the computation time to download 20 1-degree tiles using several cores.
elapsed_time = (end_time - start_time) / 60
print('Execution time:', round(elapsed_time, 2), 'minutes')
Execution time: 9.1 minutes
Transform multiband fcc raster in one band raster¶
We transform the data to have only one band describing the forest cover change with 0 for non-forest, 1 for deforestation on the period 2001–2009, 2 for deforestation on the period 2010–2019, and 3 for the remaining forest in 2020. To do so, we just sum the values of the raster bands.
sum_raster_bands(input_file="out_gfc80/forest_gfc80.tif",
output_file="out_gfc80/fcc_gfc80.tif",
verbose=False)
We resample at a lower resolution for plotting.
infn = "out_gfc80/fcc_gfc80.tif"
outfn = "out_gfc80/fcc_gfc80_coarsen.tif"
scale = gdal.Open(infn).GetGeoTransform()[1]
xres = 20 * scale
yres = 20 * scale
resample_alg = "near"
ds = gdal.Warp(outfn, infn, xRes=xres, yRes=yres, resampleAlg=resample_alg)
ds = None
Plot the forest cover change map¶
We prepare the colors for the map.
# Colors
cols=[(255, 165, 0, 255), (227, 26, 28, 255), (34, 139, 34, 255)]
colors = [(1, 1, 1, 0)] # transparent white for 0
cmax = 255.0 # float for division
for col in cols:
col_class = tuple([i / cmax for i in col])
colors.append(col_class)
color_map = ListedColormap(colors)
# Labels
labels = {0: "non-forest in 2001", 1:"deforestation 2001-2009",
2:"deforestation 2010-2019", 3:"forest in 2020"}
patches = [mpatches.Patch(facecolor=col, edgecolor="black",
label=labels[i]) for (i, col) in enumerate(colors)]
We plot the forest cover change map.
with gdal.Open("out_gfc80/fcc_gfc80_coarsen.tif", gdal.GA_ReadOnly) as ds:
raster_image = ds.ReadAsArray()
nrow, ncol = raster_image.shape
xmin, xres, _, ymax, _, yres = ds.GetGeoTransform()
extent = [xmin, xmin + xres * ncol, ymax + yres * nrow, ymax]
# Plot
fig = plt.figure()
ax = plt.subplot(111)
ax.imshow(raster_image, cmap=color_map, extent=extent,
resample=False)
ax.set_aspect("equal")
grid_image = grid.boundary.plot(ax=ax, color="grey", linewidth=0.5)
borders_image = borders.boundary.plot(ax=ax, color="black", linewidth=0.5)
plt.title("Forest cover change 2001-2010-2020, GFC 80%")
plt.legend(handles=patches, bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.xlim((163, 169))
fig.savefig("fcc_gfc80.png", bbox_inches="tight", dpi=200)
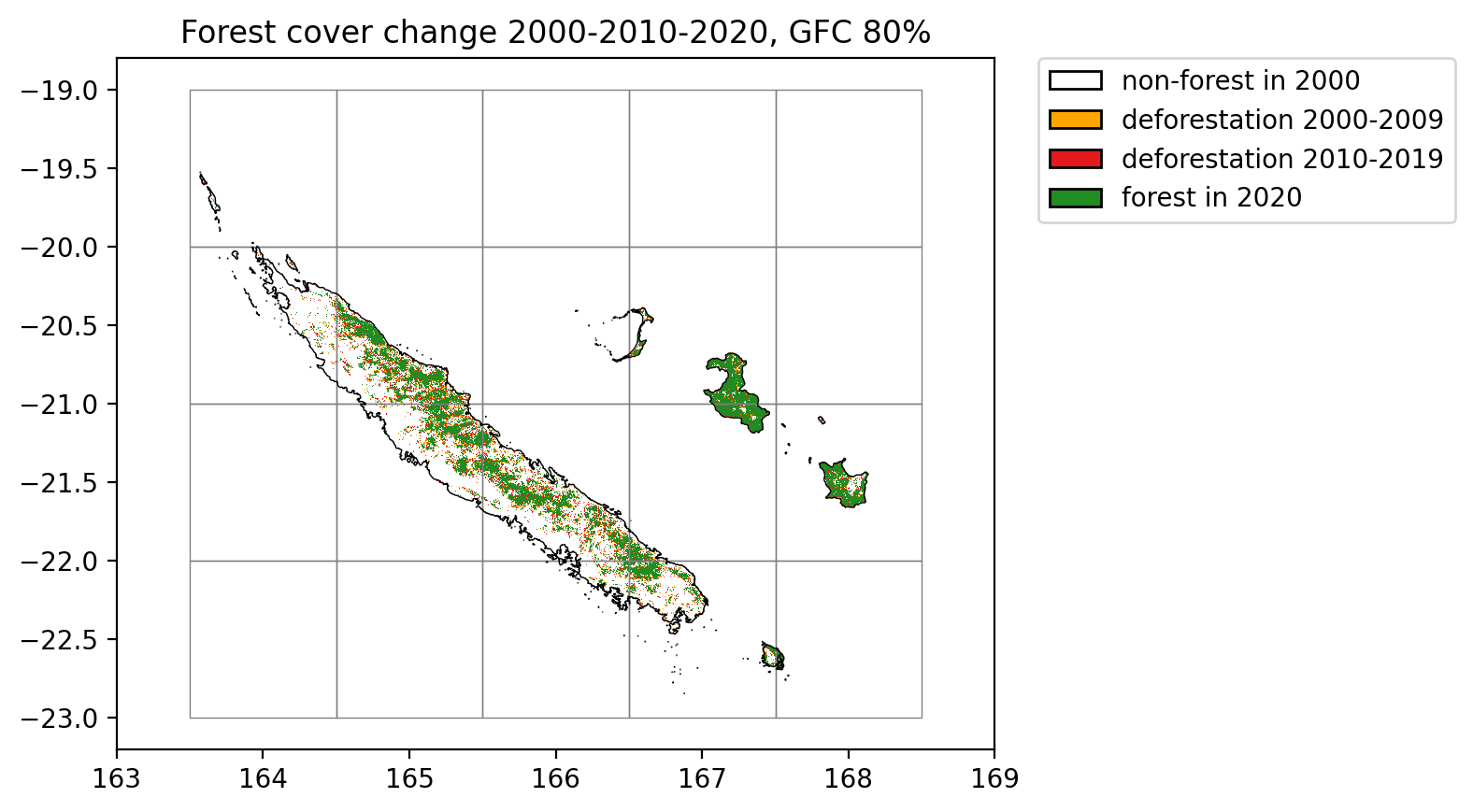
Lines in black represent country borders and the 10 km buffer. One degree tiles in grey cover the whole buffer and were used to download the data in parallel.
Reproject in EPSG:3163 for area computation¶
ifile = os.path.join("out_gfc80", "fcc_gfc80.tif")
ofile = os.path.join("out_gfc80", "fcc_gfc80_epsg3163.tif")
ds = gdal.Warp(ofile, ifile, xRes=30, yRes=30, dstSRS="EPSG:3163", resampleAlg="near",
targetAlignedPixels=True, creationOptions=["COMPRESS=DEFLATE"])
ds = None
Compute statistics¶
pixel_count = [n1, n2, n3] = [41629, 27698, 7276643]
areas = [round(i * (30 * 30 / 10000)) for i in pixel_count]
gfc80_areas= {"product": "gfc", "version": "v1_11(2023)", "perc": 80,
"fc2001": areas[0] + areas[1] + areas[2],
"fc2010": areas[1] + areas[2], "fc2020": areas[2],
"d1": round(areas[0] / 9), "d2": round(areas[1] / 10)}
print(gfc80_areas)
{'product': 'gfc', 'version': 'v1_11(2023)', 'perc': 80, 'fc2001': 661138, 'fc2010': 657391, 'fc2020': 654898, 'd1': 416, 'd2': 249}
Using GFC product and tree cover \(\ge\) 60%¶
Downloading data¶
We download the forest cover change data from GEE for New Caledonia for years 2000, 2010 and 2020, using a tile size of one degree. We use the GFC product (version v1_11 available in geefcc) and a tree cover percentage \(\ge\) 60 to define the forest.
start_time = time.time()
get_fcc(
aoi=(163.5, -23, 168.15, -19.51),
buff=0.0,
years=[2001, 2010, 2020],
source="gfc",
perc=60,
tile_size=1.0,
crop_to_aoi=True,
output_file="out_gfc60/forest_gfc60.tif",
)
end_time = time.time()
We estimate the computation time to download 20 1-degree tiles using several cores.
elapsed_time = (end_time - start_time) / 60
print('Execution time:', round(elapsed_time, 2), 'minutes')
Execution time: 8.87 minutes
Transform multiband fcc raster in one band raster¶
We transform the data to have only one band describing the forest cover change with 0 for non-forest, 1 for deforestation on the period 2001–2009, 2 for deforestation on the period 2010–2019, and 3 for the remaining forest in 2020. To do so, we just sum the values of the raster bands.
sum_raster_bands(input_file="out_gfc60/forest_gfc60.tif",
output_file="out_gfc60/fcc_gfc60.tif",
verbose=False)
We resample at a lower resolution for plotting.
infn = "out_gfc60/fcc_gfc60.tif"
outfn = "out_gfc60/fcc_gfc60_coarsen.tif"
scale = gdal.Open(infn).GetGeoTransform()[1]
xres = 20 * scale
yres = 20 * scale
resample_alg = "near"
ds = gdal.Warp(outfn, infn, xRes=xres, yRes=yres, resampleAlg=resample_alg)
ds = None
Plot the forest cover change map¶
We plot the forest cover change map.
with gdal.Open("out_gfc60/fcc_gfc60_coarsen.tif", gdal.GA_ReadOnly) as ds:
raster_image = ds.ReadAsArray()
nrow, ncol = raster_image.shape
xmin, xres, _, ymax, _, yres = ds.GetGeoTransform()
extent = [xmin, xmin + xres * ncol, ymax + yres * nrow, ymax]
# Plot
fig = plt.figure()
ax = plt.subplot(111)
ax.imshow(raster_image, cmap=color_map, extent=extent,
resample=False)
ax.set_aspect("equal")
grid_image = grid.boundary.plot(ax=ax, color="grey", linewidth=0.5)
borders_image = borders.boundary.plot(ax=ax, color="black", linewidth=0.5)
plt.title("Forest cover change 2001-2010-2020, GFC 50%")
plt.legend(handles=patches, bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.xlim((163, 169))
fig.savefig("fcc_gfc60.png", bbox_inches="tight", dpi=200)
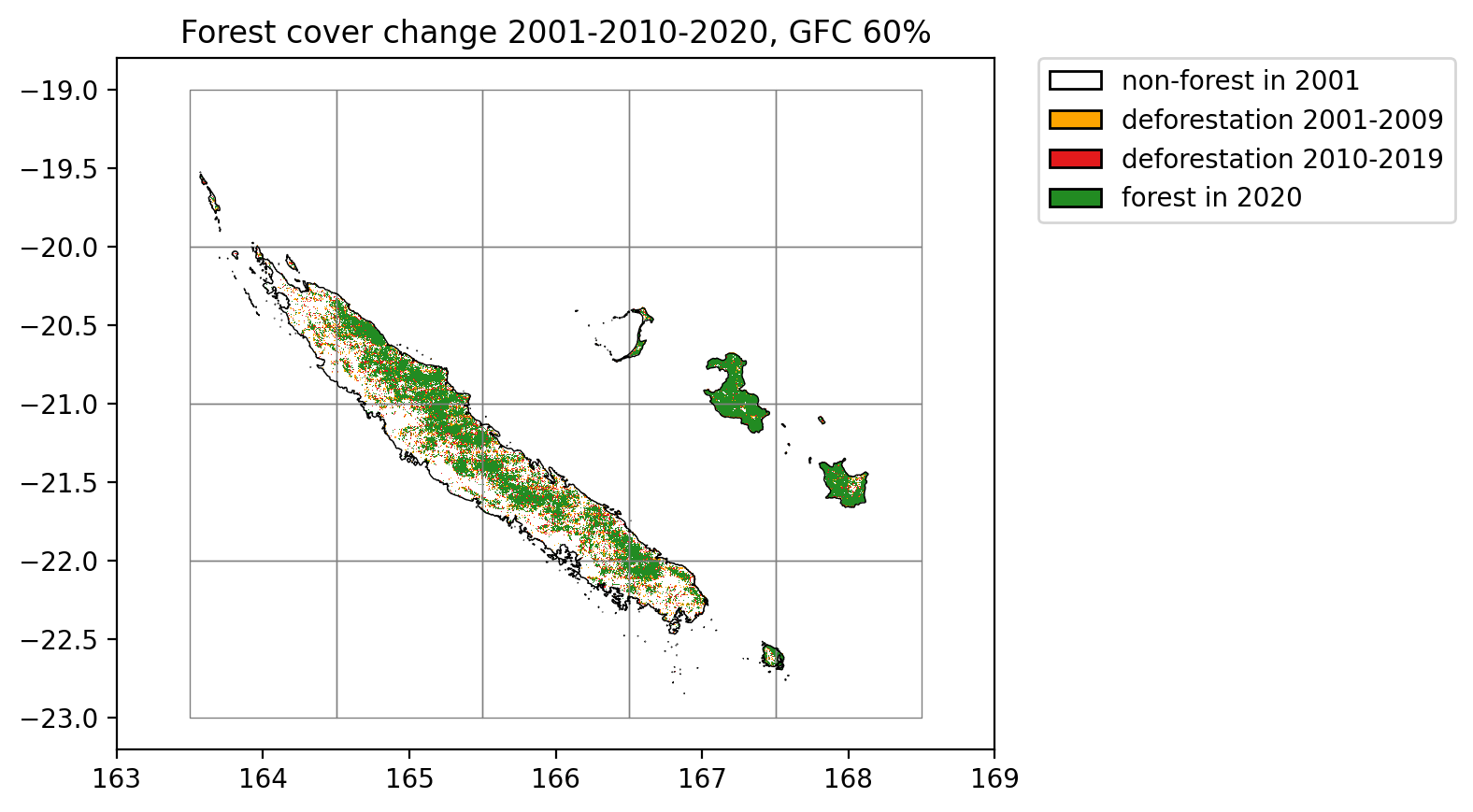
Lines in black represent country borders and the 10 km buffer. One degree tiles in grey cover the whole buffer and were used to download the data in parallel.
Reproject in EPSG:3163 for area computation¶
ifile = os.path.join("out_gfc60", "fcc_gfc60.tif")
ofile = os.path.join("out_gfc60", "fcc_gfc60_epsg3163.tif")
ds = gdal.Warp(ofile, ifile, xRes=30, yRes=30, dstSRS="EPSG:3163", resampleAlg="near",
targetAlignedPixels=True, creationOptions=["COMPRESS=DEFLATE"])
ds = None
Compute statistics¶
pixel_count = [n1, n2, n3] = [73891, 60240, 9860795]
areas = [round(i * (30 * 30 / 10000)) for i in pixel_count]
gfc60_areas= {"product": "gfc", "version": "v1_11(2023)", "perc": 60,
"fc2001": areas[0] + areas[1] + areas[2],
"fc2010": areas[1] + areas[2], "fc2020": areas[2],
"d1": round(areas[0] / 9), "d2": round(areas[1] / 10)}
print(gfc60_areas)
{'product': 'gfc', 'version': 'v1_11(2023)', 'perc': 60, 'fc2001': 899544, 'fc2010': 892894, 'fc2020': 887472, 'd1': 739, 'd2': 542}
Summary of the results¶
res_df = pd.DataFrame([tmf_areas, gfc80_areas, gfc60_areas])
res_df.to_csv(os.path.join("comparison_geefcc_nc.csv"), index=False)
tabulate(res_df, headers=res_df.columns, tablefmt="orgtbl")
product |
version |
perc |
fc2001 |
fc2010 |
fc2020 |
d1 |
d2 |
|
|---|---|---|---|---|---|---|---|---|
0 |
tmf |
v1_2023 |
893964 |
869352 |
846182 |
2735 |
2317 |
|
1 |
gfc |
v1_11(2023) |
80 |
661138 |
657391 |
654898 |
416 |
249 |
2 |
gfc |
v1_11(2023) |
60 |
899544 |
892894 |
887472 |
739 |
542 |
Forest cover for TMF and GFC with tree cover \(\ge\) 60% are similar in 2020 (about 850,000 ha) but the annual deforestation is 4-5 times lower when using the GFC product (e.g. 542 ha/yr for GFC in the period 2010–2020 against 2317 ha/yr for TMF for the same period).