ForestAtRisk Tropics¶
This notebook provides a minimal and reproducible example for the following scientific article:
Vieilledent G., C. Vancutsem, C. Bourgoin, P. Ploton, P. Verley, and F. Achard. Spatial scenario of tropical deforestation and carbon emissions for the 21st century. bioRxiv. [doi:10.1101/2022.03.22.485306]. Manuscript. Supplementary Information.
We use the Guadeloupe archipelago as a case study.
[1]:
# Imports
import os
import re
from shutil import copy2
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from patsy import dmatrices
import pickle
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import log_loss
import forestatrisk as far
# forestatrisk: modelling and forecasting deforestation in the tropics.
# https://ecology.ghislainv.fr/forestatrisk/
We create a directory to hold the outputs with the help of the function .make_dir().
[2]:
# Make output directory
far.make_dir("output")
1. Data¶
1.1 Import and unzip the data¶
[3]:
import urllib.request
from zipfile import ZipFile
# Source of the data
url = "https://github.com/ghislainv/forestatrisk/raw/master/docsrc/notebooks/data_GLP.zip"
if os.path.exists("data_GPL.zip") is False:
urllib.request.urlretrieve(url, "data_GPL.zip")
with ZipFile("data_GPL.zip", "r") as z:
z.extractall("data")
1.2 Files¶
The data folder includes:
Forest cover change data for the period 2010-2020 as a GeoTiff raster file (
data/fcc23.tif).Spatial explanatory variables as GeoTiff raster files (
.tifextension, eg.data/dist_edge.tiffor distance to forest edge).Additional folders:
forest,forecast, andemissions, with forest cover change for different periods of time, explanatory variables at different dates used for projections in the future, and forest carbon data for computing carbon emissions.
[4]:
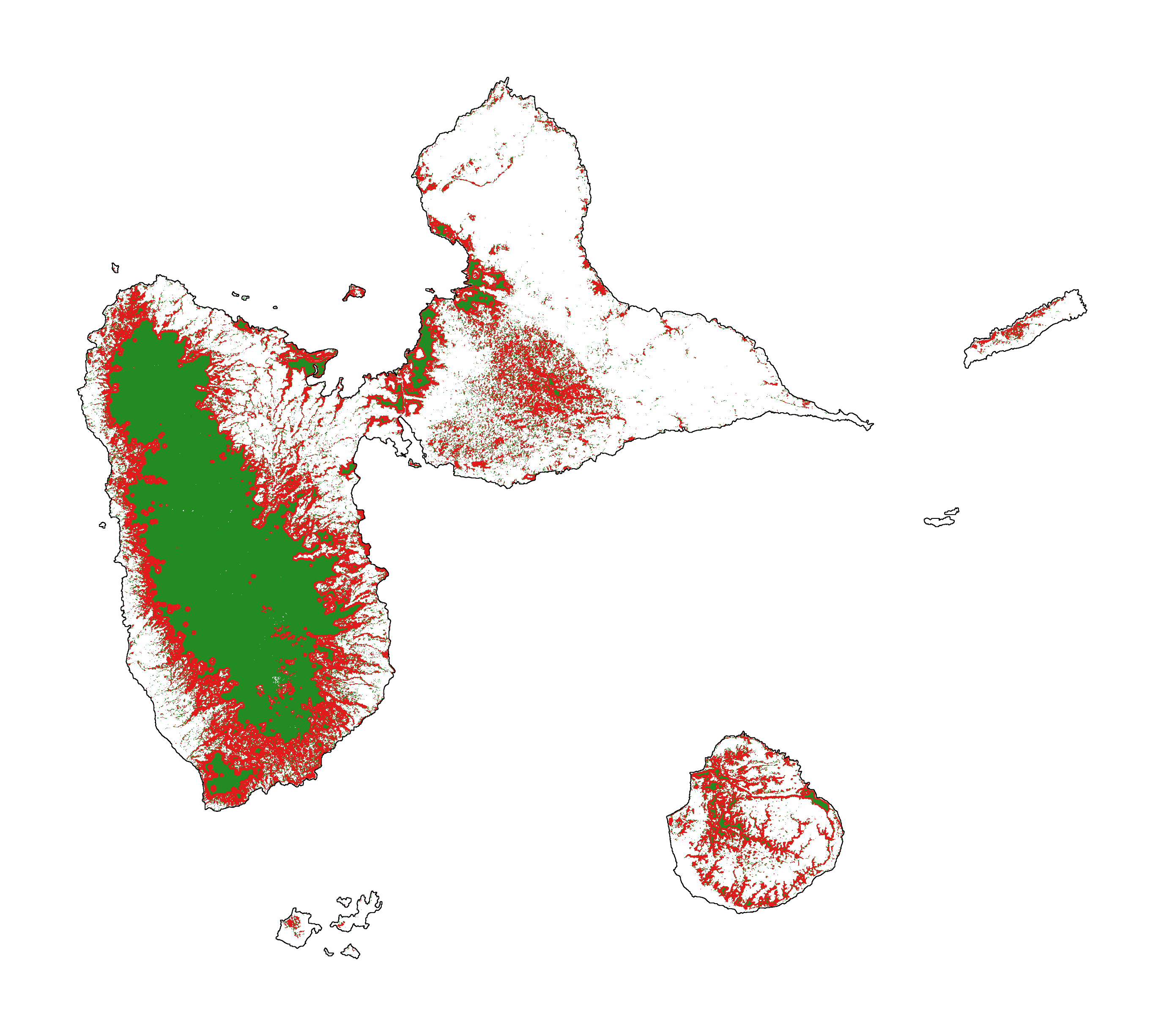
# Plot forest
fig_fcc23 = far.plot.fcc(
input_fcc_raster="data/fcc23.tif",
maxpixels=1e8,
output_file="output/fcc23.png",
borders="data/aoi_proj.shp",
linewidth=0.3, dpi=500)
1.3 Sampling the observations¶
[5]:
# Sample points
dataset = far.sample(nsamp=10000, adapt=True, seed=1234, csize=10,
var_dir="data",
input_forest_raster="fcc23.tif",
output_file="output/sample.txt",
blk_rows=0)
Sample 2x 10000 pixels (deforested vs. forest)
Divide region in 168 blocks
Compute number of deforested and forest pixels per block
100%
Draw blocks at random
Draw pixels at random in blocks
100%
Compute center of pixel coordinates
Compute number of 10 x 10 km spatial cells
... 99 cells (9 x 11)
Identify cell number from XY coordinates
Make virtual raster with variables as raster bands
Extract raster values for selected pixels
100%
Export results to file output/sample.txt
[6]:
# Remove NA from data-set (otherwise scale() and
# model_binomial_iCAR doesn't work)
dataset = dataset.dropna(axis=0)
# Set number of trials to one for far.model_binomial_iCAR()
dataset["trial"] = 1
# Print the first five rows
print(dataset.head(5))
altitude dist_defor dist_edge dist_river dist_road dist_town fcc23 \
0 30.0 642.0 30.0 8448.0 1485.0 6364.0 0.0
1 37.0 765.0 30.0 8583.0 1697.0 6576.0 0.0
2 78.0 216.0 30.0 7722.0 949.0 5743.0 0.0
3 80.0 277.0 30.0 8168.0 1172.0 6047.0 0.0
4 46.0 30.0 30.0 6179.0 541.0 6690.0 0.0
pa slope X Y cell trial
0 0.0 8.0 -6842295.0 1851975.0 4.0 1
1 0.0 7.0 -6842235.0 1852095.0 4.0 1
2 0.0 5.0 -6842535.0 1851195.0 4.0 1
3 0.0 2.0 -6842445.0 1851615.0 4.0 1
4 0.0 1.0 -6840465.0 1849755.0 4.0 1
[7]:
# Sample size
ndefor = sum(dataset.fcc23 == 0)
nfor = sum(dataset.fcc23 == 1)
with open("output/sample_size.csv", "w") as f:
f.write("var, n\n")
f.write("ndefor, " + str(ndefor) + "\n")
f.write("nfor, " + str(nfor) + "\n")
print("ndefor = {}, nfor = {}".format(ndefor, nfor))
ndefor = 9923, nfor = 9979
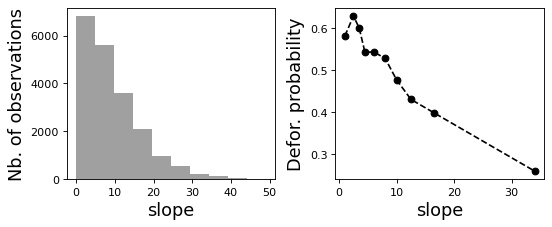
1.4 Correlation plots¶
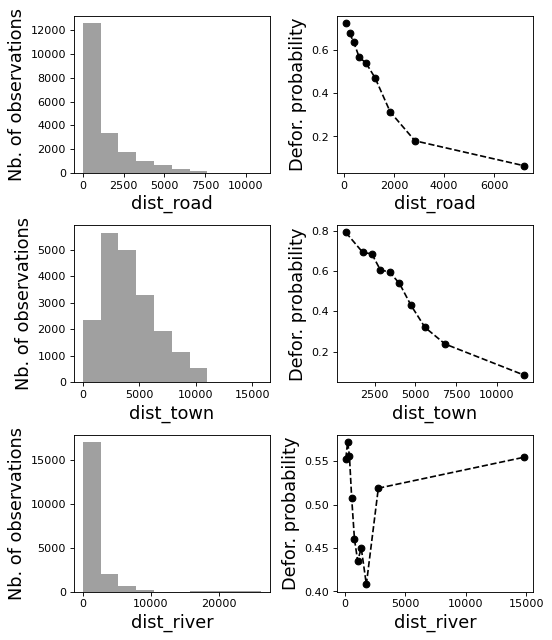
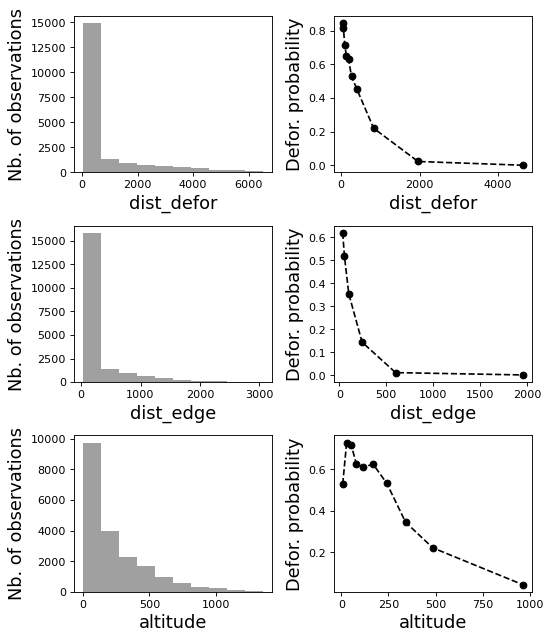
[8]:
# Correlation formula
formula_corr = "fcc23 ~ dist_road + dist_town + dist_river + \
dist_defor + dist_edge + altitude + slope - 1"
# Output file
of = "output/correlation.pdf"
# Data
y, data = dmatrices(formula_corr, data=dataset,
return_type="dataframe")
# Plots
figs = far.plot.correlation(
y=y, data=data,
plots_per_page=3,
figsize=(7, 8),
dpi=80,
output_file=of)
2. Model¶
2.1 Model preparation¶
[9]:
# Neighborhood for spatial-autocorrelation
nneigh, adj = far.cellneigh(raster="data/fcc23.tif", csize=10, rank=1)
# List of variables
variables = ["C(pa)", "scale(altitude)", "scale(slope)",
"scale(dist_defor)", "scale(dist_edge)", "scale(dist_road)",
"scale(dist_town)", "scale(dist_river)"]
# Transform into numpy array
variables = np.array(variables)
# Starting values
beta_start = -99 # Simple GLM estimates
# Priors
priorVrho = -1 # -1="1/Gamma"
Compute number of 10 x 10 km spatial cells
... 99 cells (9 x 11)
Identify adjacent cells and compute number of neighbors
2.2 Variable selection¶
[10]:
# Run model while there is non-significant variables
var_remove = True
while(np.any(var_remove)):
# Formula
right_part = " + ".join(variables) + " + cell"
left_part = "I(1-fcc23) + trial ~ "
formula = left_part + right_part
# Model
mod_binomial_iCAR = far.model_binomial_iCAR(
# Observations
suitability_formula=formula, data=dataset,
# Spatial structure
n_neighbors=nneigh, neighbors=adj,
# Priors
priorVrho=priorVrho,
# Chains
burnin=1000, mcmc=1000, thin=1,
# Starting values
beta_start=beta_start)
# Ecological and statistical significance
effects = mod_binomial_iCAR.betas[1:]
# MCMC = mod_binomial_iCAR.mcmc
# CI_low = np.percentile(MCMC, 2.5, axis=0)[1:-2]
# CI_high = np.percentile(MCMC, 97.5, axis=0)[1:-2]
positive_effects = (effects >= 0)
# zero_in_CI = ((CI_low * CI_high) <= 0)
# Keeping only significant variables
var_remove = positive_effects
# var_remove = np.logical_or(positive_effects, zero_in_CI)
var_keep = np.logical_not(var_remove)
variables = variables[var_keep]
Using estimates from classic logistic regression as starting values for betas
Using estimates from classic logistic regression as starting values for betas
2.3 Final model¶
[11]:
# Re-run the model with longer MCMC and estimated initial values
mod_binomial_iCAR = far.model_binomial_iCAR(
# Observations
suitability_formula=formula, data=dataset,
# Spatial structure
n_neighbors=nneigh, neighbors=adj,
# Priors
priorVrho=priorVrho,
# Chains
burnin=5000, mcmc=5000, thin=5,
# Starting values
beta_start=mod_binomial_iCAR.betas)
2.4 Model summary¶
[12]:
# Predictions
pred_icar = mod_binomial_iCAR.theta_pred
# Summary
print(mod_binomial_iCAR)
# Write summary in file
with open("output/summary_hSDM.txt", "w") as f:
f.write(str(mod_binomial_iCAR))
Binomial logistic regression with iCAR process
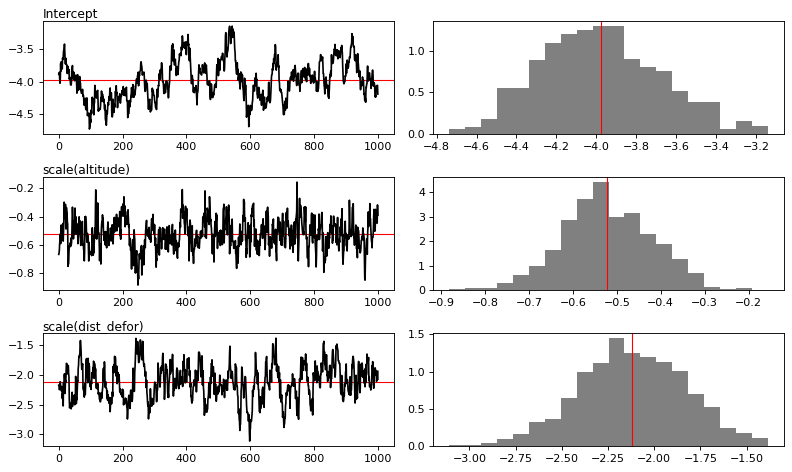
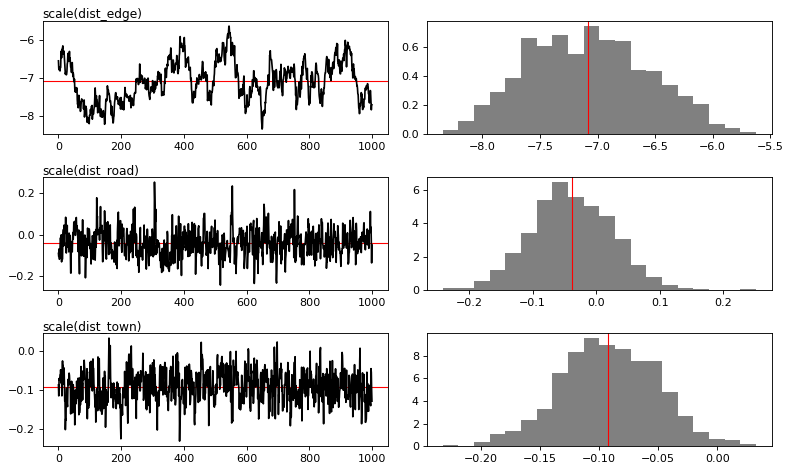
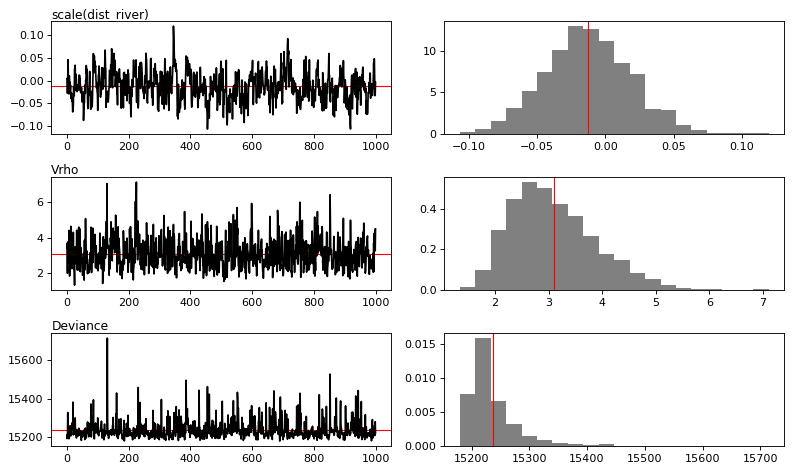
Model: I(1 - fcc23) + trial ~ 1 + scale(altitude) + scale(dist_defor) + scale(dist_edge) + scale(dist_road) + scale(dist_town) + scale(dist_river) + cell
Posteriors:
Mean Std CI_low CI_high
Intercept -3.98 0.303 -4.5 -3.37
scale(altitude) -0.522 0.105 -0.731 -0.321
scale(dist_defor) -2.12 0.288 -2.71 -1.56
scale(dist_edge) -7.08 0.525 -8.01 -6.08
scale(dist_road) -0.0389 0.0657 -0.162 0.0885
scale(dist_town) -0.092 0.041 -0.176 -0.0134
scale(dist_river) -0.0129 0.0319 -0.0735 0.0502
Vrho 3.1 0.815 1.86 4.92
Deviance 1.52e+04 48.1 1.52e+04 1.54e+04
MCMC traces
[13]:
# Plot
figs = mod_binomial_iCAR.plot(
output_file="output/mcmc.pdf",plots_per_page=3,
figsize=(10, 6),
dpi=80)
Traces and posteriors will be plotted in output/mcmc.pdf
[14]:
# Save model's main specifications with pickle
mod_icar_pickle = {
"formula": mod_binomial_iCAR.suitability_formula,
"rho": mod_binomial_iCAR.rho,
"betas": mod_binomial_iCAR.betas,
"Vrho": mod_binomial_iCAR.Vrho,
"deviance": mod_binomial_iCAR.deviance}
with open("output/mod_icar.pickle", "wb") as pickle_file:
pickle.dump(mod_icar_pickle, pickle_file)
3. Model comparison and validation¶
3.1 Cross-validation¶
[15]:
# Cross-validation for icar, glm and RF
CV_df_icar = far.cross_validation(
dataset, formula, mod_type="icar", ratio=30, nrep=5,
icar_args={"n_neighbors": nneigh, "neighbors": adj,
"burnin": 1000, "mcmc": 1000, "thin": 1,
"beta_start": mod_binomial_iCAR.betas})
CV_df_glm = far.cross_validation(dataset, formula, mod_type="glm", ratio=30, nrep=5)
CV_df_rf = far.cross_validation(dataset, formula, mod_type="rf", ratio=30, nrep=5,
rf_args={"n_estimators": 500, "n_jobs": 3})
# Save result to disk
CV_df_icar.to_csv("output/CV_icar.csv", header=True, index=False)
CV_df_glm.to_csv("output/CV_glm.csv", header=True, index=False)
CV_df_rf.to_csv("output/CV_rf.csv", header=True, index=False)
Repetition #: 1
Repetition #: 2
Repetition #: 3
Repetition #: 4
Repetition #: 5
Repetition #: 1
Repetition #: 2
Repetition #: 3
Repetition #: 4
Repetition #: 5
Repetition #: 1
Repetition #: 2
Repetition #: 3
Repetition #: 4
Repetition #: 5
[16]:
print(CV_df_icar)
index rep1 rep2 rep3 rep4 rep5 mean
0 AUC 0.9070 0.9065 0.9040 0.9032 0.9037 0.9049
1 OA 0.8245 0.8245 0.8218 0.8191 0.8164 0.8213
2 EA 0.5001 0.5002 0.5001 0.5000 0.5000 0.5001
3 FOM 0.7044 0.6962 0.6946 0.6952 0.6875 0.6956
4 Sen 0.8265 0.8209 0.8198 0.8202 0.8148 0.8204
5 Spe 0.8224 0.8279 0.8238 0.8181 0.8181 0.8220
6 TSS 0.6489 0.6488 0.6436 0.6382 0.6329 0.6425
7 K 0.6489 0.6488 0.6436 0.6382 0.6329 0.6425
[17]:
print(CV_df_glm)
index rep1 rep2 rep3 rep4 rep5 mean
0 AUC 0.8843 0.8834 0.8810 0.8842 0.8877 0.8841
1 OA 0.8037 0.7980 0.7937 0.8017 0.8041 0.8002
2 EA 0.5001 0.5002 0.5001 0.5000 0.5000 0.5001
3 FOM 0.6751 0.6583 0.6547 0.6707 0.6699 0.6657
4 Sen 0.8060 0.7939 0.7913 0.8029 0.8023 0.7993
5 Spe 0.8014 0.8020 0.7960 0.8005 0.8058 0.8011
6 TSS 0.6074 0.5959 0.5873 0.6034 0.6081 0.6004
7 K 0.6074 0.5959 0.5873 0.6034 0.6081 0.6004
[18]:
print(CV_df_rf)
index rep1 rep2 rep3 rep4 rep5 mean
0 AUC 0.9133 0.9063 0.9116 0.9100 0.9107 0.9104
1 OA 0.8339 0.8235 0.8292 0.8280 0.8282 0.8285
2 EA 0.5001 0.5001 0.5000 0.5000 0.5000 0.5001
3 FOM 0.7175 0.6956 0.7099 0.7089 0.7056 0.7075
4 Sen 0.8369 0.8216 0.8303 0.8298 0.8279 0.8293
5 Spe 0.8307 0.8253 0.8280 0.8262 0.8284 0.8277
6 TSS 0.6677 0.6469 0.6583 0.6560 0.6563 0.6570
7 K 0.6677 0.6469 0.6583 0.6560 0.6563 0.6570
3.2 Deviance¶
[19]:
# Null model
formula_null = "I(1-fcc23) ~ 1"
y, x = dmatrices(formula_null, data=dataset, NA_action="drop")
Y = y[:, 0]
X_null = x[:, :]
mod_null = LogisticRegression(solver="lbfgs")
mod_null = mod_null.fit(X_null, Y)
pred_null = mod_null.predict_proba(X_null)
# Simple glm with no spatial random effects
formula_glm = formula
y, x = dmatrices(formula_glm, data=dataset, NA_action="drop")
Y = y[:, 0]
X_glm = x[:, :-1] # We remove the last column (cells)
mod_glm = LogisticRegression(solver="lbfgs")
mod_glm = mod_glm.fit(X_glm, Y)
pred_glm = mod_glm.predict_proba(X_glm)
# Random forest model
formula_rf = formula
y, x = dmatrices(formula_rf, data=dataset, NA_action="drop")
Y = y[:, 0]
X_rf = x[:, :-1] # We remove the last column (cells)
mod_rf = RandomForestClassifier(n_estimators=500, n_jobs=3)
mod_rf = mod_rf.fit(X_rf, Y)
pred_rf = mod_rf.predict_proba(X_rf)
# Deviances
deviance_null = 2*log_loss(Y, pred_null, normalize=False)
deviance_glm = 2*log_loss(Y, pred_glm, normalize=False)
deviance_rf = 2*log_loss(Y, pred_rf, normalize=False)
deviance_icar = mod_binomial_iCAR.deviance
deviance_full = 0
dev = [deviance_null, deviance_glm, deviance_rf, deviance_icar, deviance_full]
# Result table
mod_dev = pd.DataFrame({"model": ["null", "glm", "rf", "icar", "full"],
"deviance": dev})
perc = 100*(1-mod_dev.deviance/deviance_null)
mod_dev["perc"] = perc
mod_dev = mod_dev.round(0)
mod_dev.to_csv("output/model_deviance.csv", header=True, index=False)
[20]:
print(mod_dev)
model deviance perc
0 null 27590.0 0.0
1 glm 16542.0 40.0
2 rf 3646.0 87.0
3 icar 15237.0 45.0
4 full 0.0 100.0
[21]:
# Save models' predictions
obs_pred = dataset
obs_pred["null"] = pred_null[:, 1]
obs_pred["glm"] = pred_glm[:, 1]
obs_pred["rf"] = pred_rf[:, 1]
obs_pred["icar"] = pred_icar
obs_pred.to_csv("output/obs_pred.csv", header=True, index=False)
4. Predict¶
4.1 Interpolate spatial random effects¶
[22]:
# Spatial random effects
rho = mod_binomial_iCAR.rho
# Interpolate
far.interpolate_rho(rho=rho, input_raster="data/fcc23.tif",
output_file="output/rho.tif",
csize_orig=10, csize_new=1)
Write spatial random effect data to disk
Compute statistics
Build overview
Resampling spatial random effects to file output/rho.tif
4.2 Predict deforestation probability¶
[23]:
# Update dist_edge and dist_defor at t3
os.rename("data/dist_edge.tif", "data/dist_edge.tif.bak")
os.rename("data/dist_defor.tif", "data/dist_defor.tif.bak")
copy2("data/forecast/dist_edge_forecast.tif", "data/dist_edge.tif")
copy2("data/forecast/dist_defor_forecast.tif", "data/dist_defor.tif")
# Compute predictions
far.predict_raster_binomial_iCAR(
mod_binomial_iCAR, var_dir="data",
input_cell_raster="output/rho.tif",
input_forest_raster="data/forest/forest_t3.tif",
output_file="output/prob.tif",
blk_rows=10 # Reduced number of lines to avoid memory problems
)
# Reinitialize data
os.remove("data/dist_edge.tif")
os.remove("data/dist_defor.tif")
os.rename("data/dist_edge.tif.bak", "data/dist_edge.tif")
os.rename("data/dist_defor.tif.bak", "data/dist_defor.tif")
Make virtual raster with variables as raster bands
Divide region in 296 blocks
Create a raster file on disk for projections
Predict deforestation probability by block
100%
Compute statistics
5. Project future forest cover change¶
[24]:
# Forest cover
fc = list()
dates = ["t1", "2005", "t2", "2015", "t3"]
ndates = len(dates)
for i in range(ndates):
rast = "data/forest/forest_" + dates[i] + ".tif"
val = far.countpix(input_raster=rast, value=1)
fc.append(val["area"]) # area in ha
# Save results to disk
f = open("output/forest_cover.txt", "w")
for i in fc:
f.write(str(i) + "\n")
f.close()
# Annual deforestation
T = 10.0
annual_defor = (fc[2] - fc[4]) / T
# Dates and time intervals
dates_fut = ["2030", "2035", "2040", "2050", "2055", "2060", "2070", "2080", "2085", "2090", "2100"]
ndates_fut = len(dates_fut)
ti = [10, 15, 20, 30, 35, 40, 50, 60, 65, 70, 80]
Divide region in 168 blocks
Compute the number of pixels with value=1
100%
Compute the corresponding area in ha
Divide region in 168 blocks
Compute the number of pixels with value=1
100%
Compute the corresponding area in ha
Divide region in 168 blocks
Compute the number of pixels with value=1
100%
Compute the corresponding area in ha
Divide region in 168 blocks
Compute the number of pixels with value=1
100%
Compute the corresponding area in ha
Divide region in 168 blocks
Compute the number of pixels with value=1
100%
Compute the corresponding area in ha
[25]:
# Loop on dates
for i in range(ndates_fut):
# Amount of deforestation (ha)
defor = np.rint(annual_defor * ti[i])
# Compute future forest cover
stats = far.deforest(
input_raster="output/prob.tif",
hectares=defor,
output_file="output/fcc_" + dates_fut[i] + ".tif",
blk_rows=128)
# Save some stats if date = 2050
if dates_fut[i] == "2050":
# Save stats to disk with pickle
pickle.dump(stats, open("output/stats.pickle", "wb"))
# Plot histograms of probabilities
fig_freq = far.plot.freq_prob(
stats, output_file="output/freq_prob.png")
plt.close(fig_freq)
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
Divide region in 24 blocks
Compute the total number of forest pixels
100%
Identify threshold
Minimize error on deforested hectares
Create a raster file on disk for forest-cover change
Write raster of future forest-cover change
100%
Compute statistics
6. Carbon emissions¶
[26]:
# Create dataframe
dpast = ["2020"]
dpast.extend(dates_fut)
C_df = pd.DataFrame({"date": dpast, "C": np.repeat(-99, ndates_fut + 1)},
columns=["date","C"])
# Loop on date
for i in range(ndates_fut):
carbon = far.emissions(input_stocks="data/emissions/AGB.tif",
input_forest="output/fcc_" + dates_fut[i] + ".tif")
C_df.loc[C_df["date"]==dates_fut[i], ["C"]] = carbon
# Past emissions
carbon = far.emissions(input_stocks="data/emissions/AGB.tif",
input_forest="data/fcc23.tif")
C_df.loc[C_df["date"]==dpast[0], ["C"]] = carbon
# Save dataframe
C_df.to_csv("output/C_emissions.csv", header=True, index=False)
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
Make virtual raster
Divide region in 24 blocks
Compute carbon emissions by block
100%
[27]:
print(C_df)
date C
0 2020 85954
1 2030 98242
2 2035 151104
3 2040 201820
4 2050 289924
5 2055 334290
6 2060 381925
7 2070 481285
8 2080 587603
9 2085 644809
10 2090 705718
11 2100 843905
7. Figures¶
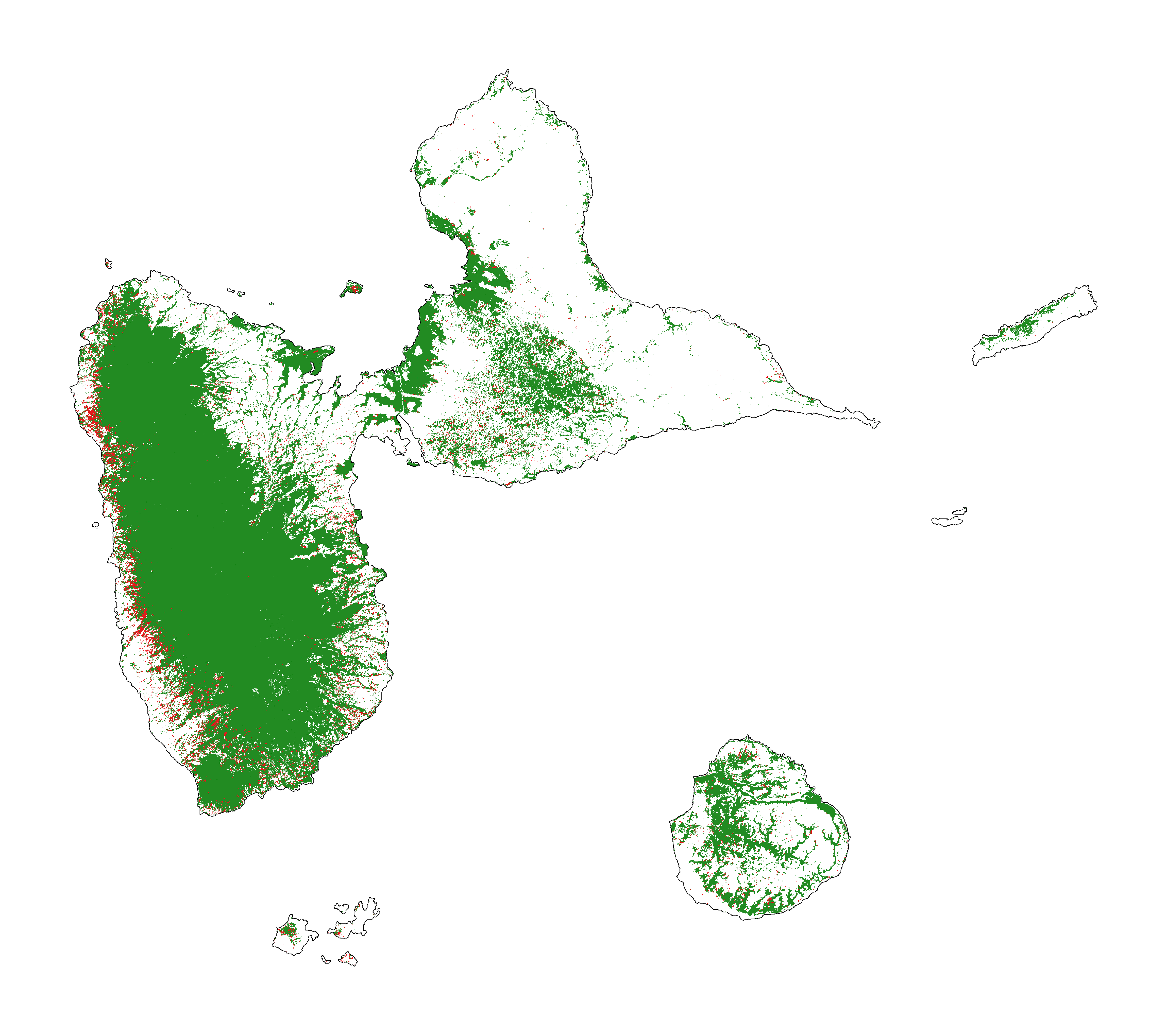
7.1 Historical forest cover change¶
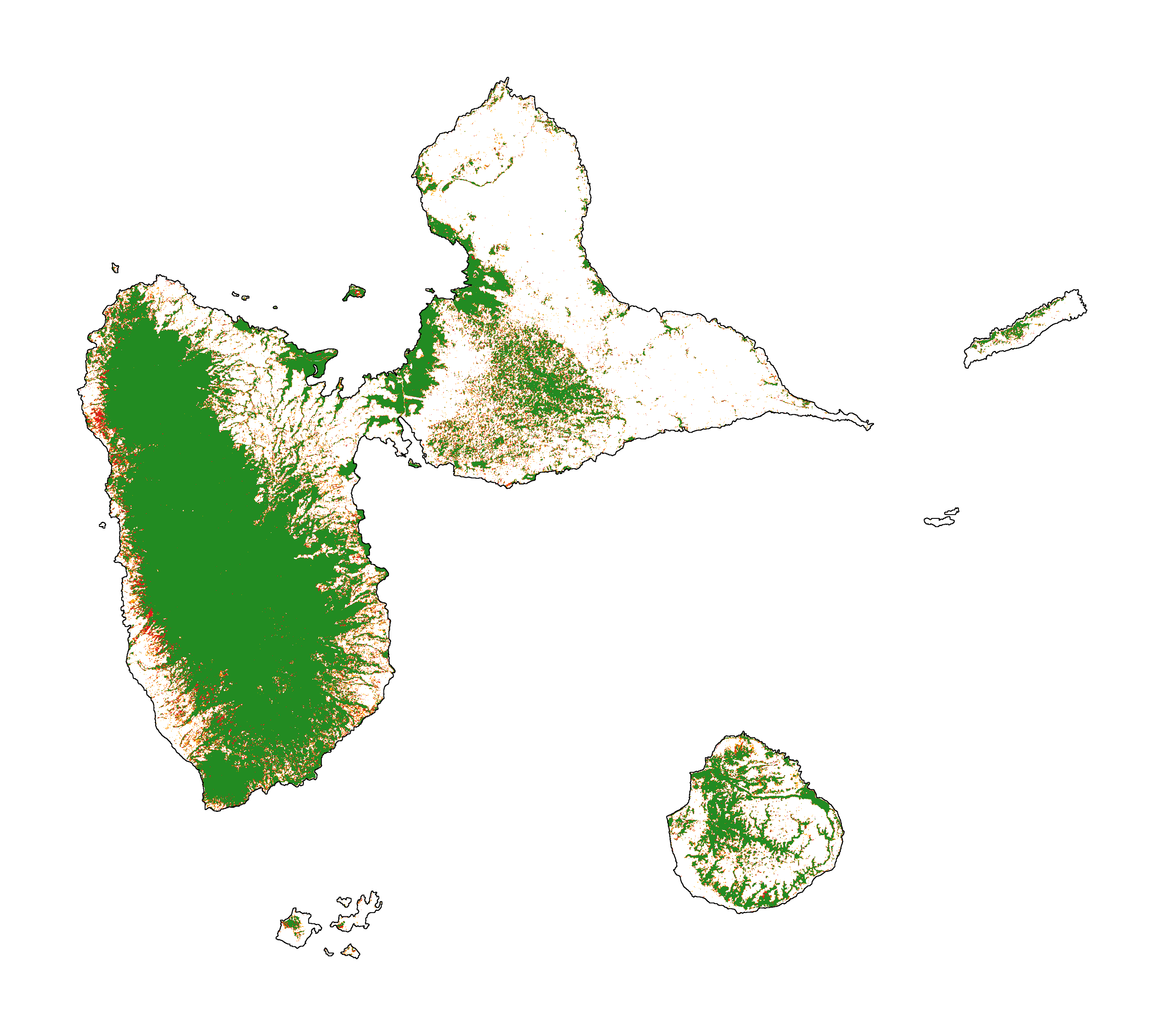
Forest cover change for the period 2000-2010-2020
[28]:
# Plot forest
fig_fcc123 = far.plot.fcc123(
input_fcc_raster="data/forest/fcc123.tif",
maxpixels=1e8,
output_file="output/fcc123.png",
borders="data/aoi_proj.shp",
linewidth=0.3,
figsize=(6, 5), dpi=500)
7.2 Spatial random effects¶
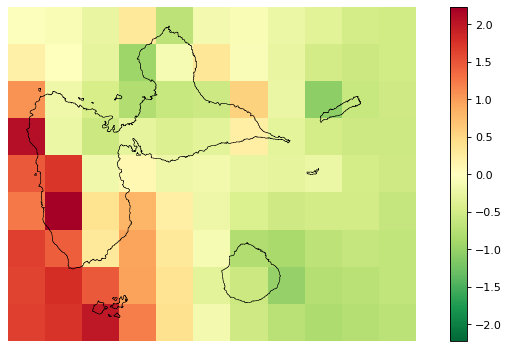
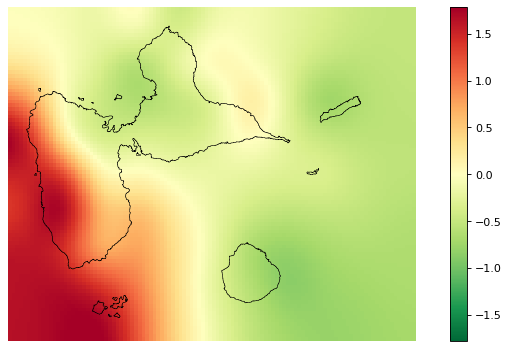
[29]:
# Original spatial random effects
fig_rho_orig = far.plot.rho("output/rho_orig.tif",
borders="data/aoi_proj.shp",
linewidth=0.5,
output_file="output/rho_orig.png",
figsize=(9,5), dpi=80)
# Interpolated spatial random effects
fig_rho = far.plot.rho("output/rho.tif",
borders="data/aoi_proj.shp",
linewidth=0.5,
output_file="output/rho.png",
figsize=(9,5), dpi=80)
Build overview
7.3 Spatial probability of deforestation¶
[30]:
# Spatial probability of deforestation
fig_prob = far.plot.prob("output/prob.tif",
maxpixels=1e8,
borders="data/aoi_proj.shp",
linewidth=0.3,
legend=True,
output_file="output/prob.png",
figsize=(6, 5), dpi=500)

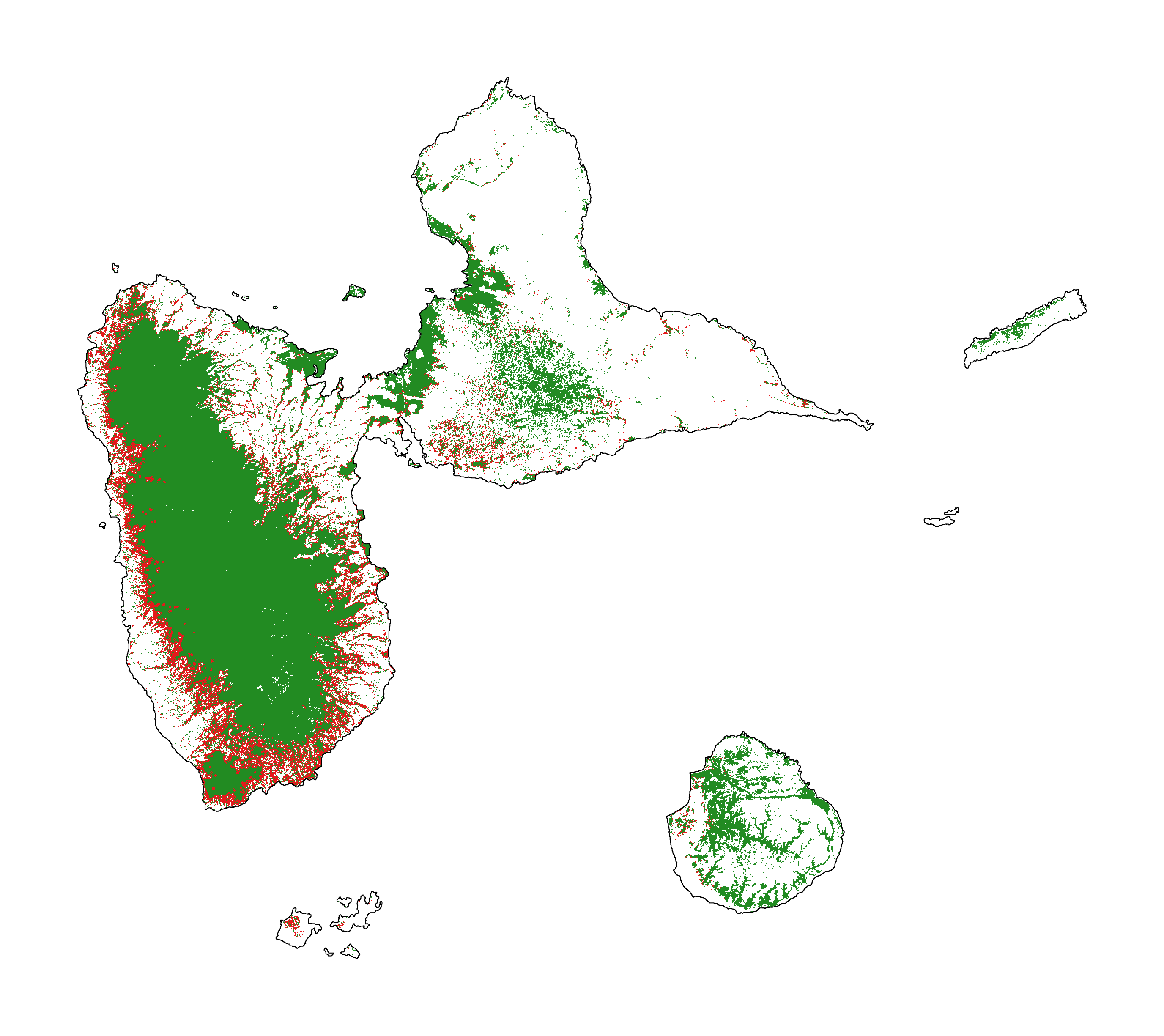
7.4 Future forest cover¶
[31]:
# Projected forest cover change (2020-2050)
fcc_2050 = far.plot.fcc("output/fcc_2050.tif",
maxpixels=1e8,
borders="data/aoi_proj.shp",
linewidth=0.3,
output_file="output/fcc_" + dates_fut[i] + ".png",
figsize=(6, 5), dpi=500)
[32]:
# Projected forest cover change (2020-2100)
fcc_2100 = far.plot.fcc("output/fcc_2100.tif",
maxpixels=1e8,
borders="data/aoi_proj.shp",
linewidth=0.3,
output_file="output/fcc_" + dates_fut[i] + ".png",
figsize=(6, 5), dpi=500)