Country data¶
Introduction¶
This notebook presents the functions of the forestatrisk Python
package that can be used to collect the spatial data needed for
modeling and forecasting deforestation in a given country.
Data are retrieved from global (or pantropical) datasets freely
available on the internet. Of course, the user can add any additional
variables to the analyses manually. To do so, the user must add
GeoTIFF raster files with extension .tif in the data folder of the
working directory.
Importing Python modules¶
We import the Python modules needed for collecting the data.
# Imports
import os
from dotenv import load_dotenv
import ee
import pandas as pd
from tabulate import tabulate
from pywdpa import get_token
import forestatrisk as far
We create some directories to hold the data and the ouputs with the
function far.make_dir().
far.make_dir("data_raw")
far.make_dir("data")
far.make_dir("output")
We increase the cache for GDAL to increase computational speed.
# GDAL
os.environ["GDAL_CACHEMAX"] = "1024"
Set credentials¶
We need to configure and set credentials for:
Google Earth Engine (GEE) API
RClone to access Google Drive
WDPA API
You will need a Google account for using the GEE API and accessing Google Drive.
Access to Google Earth Engine API¶
Google Earth Engine is used to compute the past forest cover change
from Vancutsem et al. 2021 or Hansen et al. 2013. To get credentials
for using the Google Earth Engine API, follow these
GEE instructions. While authentication with ee.Authenticate() should be
necessary only once, you have to execute the command ee.Initialize()
at each session.
# Uncomment to authenticate for the first time.
# ee.Authenticate()
ee.Initialize(project="forestatrisk")
Under Linux and Mac, credentials are stored in
$HOME/.config/earthengine/credentials.
cat $HOME/.config/earthengine/credentials
Access to Google Drive with RClone¶
RClone is used to download the forest cover change raster locally from Google Drive. To install RClone, follow these RClone instructions. To configure the access to your Google Drive, follow these RClone Google Drive instructions.
Access to WDPA API¶
We will be using the pywda Python package to collect the data on
protected areas from the World Database on Protected Areas (WDPA) at
https://www.protectedplanet.net. To access the Protected Planet API,
you must first obtain a Personal API Token by filling in the form
available at https://api.protectedplanet.net/request. Then you need to
set an environment variable (we recommend using the name WDPA_KEY)
using either the command os.environ["WDPA_KEY"]="your_token" or
python-dotenv.
The validity of your token can be checked with the function
pywdpa.get_token().
# WDPA API
load_dotenv(".env")
get_token()
If your token is valid, the function will return its value. Otherwise it will print an error message.
Data¶
Compute forest cover change¶
We specify the iso3 code of the country we want the data for, for example Martinique.
iso3 = "MTQ"
We compute the past forest cover change from Vancutsem et al. 2021
using Google Earth Engine. The argument gdrive_remote_rclone of the
function far.data.country_forest_run() specifies the name of the
Google Drive remote for rclone. The argument gdrive_folder specifies
the name of the Google Drive folder to use. In this case, folder
GEE/GEE-forestatrisk-notebooks should exist in Google Drive.
# Compute gee forest data
far.data.country_forest_run(
iso3, proj="EPSG:4326",
output_dir="data_raw",
keep_dir=True,
fcc_source="jrc", perc=50,
gdrive_remote_rclone="gdrive_gv",
gdrive_folder="GEE/GEE-forestatrisk-notebooks")
Download raw data¶
# Download data
far.data.country_download(
iso3,
gdrive_remote_rclone="gdrive_gv",
gdrive_folder="GEE/GEE-forestatrisk-notebooks",
output_dir="data_raw")
Downloading data for country MTQ
SRTM not existing for tile: 25_09
Data for MTQ have been downloaded
Compute explanatory variables¶
We first set the projection in which we want the data to be, for example EPSG:5490.
# Projection
proj = "EPSG:5490"
We compute the explanatory variables from the raw data.
# Compute variables
far.data.country_compute(
iso3,
temp_dir="data_raw",
output_dir="data",
proj=proj,
data_country=True,
data_forest=True,
keep_temp_dir=True)
Files¶
The data folder includes:
Forest cover change data for the period 2010-2020 as a GeoTiff raster file (
data/fcc23.tif).Spatial explanatory variables as GeoTiff raster files (
.tifextension, eg.data/dist_edge.tiffor distance to forest edge).Additional folders:
forest,forecast, andemissions, with forest cover change for different periods of time, explanatory variables at different dates used for projections in the future, and forest carbon data for computing carbon emissions.
Variable characteristics are summarized in the following table:
Product |
Source |
Variable |
Unit |
Resolution (m) |
|---|---|---|---|---|
Forest maps (2000-2010-2020) |
Vancutsem et al. 2021 |
distance to forest edge |
m |
30 |
distance to past deforestation |
m |
30 |
||
Digital Elevation Model |
SRTM v4.1 CSI-CGIAR |
altitude |
m |
90 |
slope |
degree |
90 |
||
Highways |
OSM-Geofabrik |
distance to roads |
m |
150 |
Places |
distance to towns |
m |
150 |
|
Waterways |
distance to river |
m |
150 |
|
Protected areas |
WDPA |
protected area presence |
– |
30 |
Plots¶
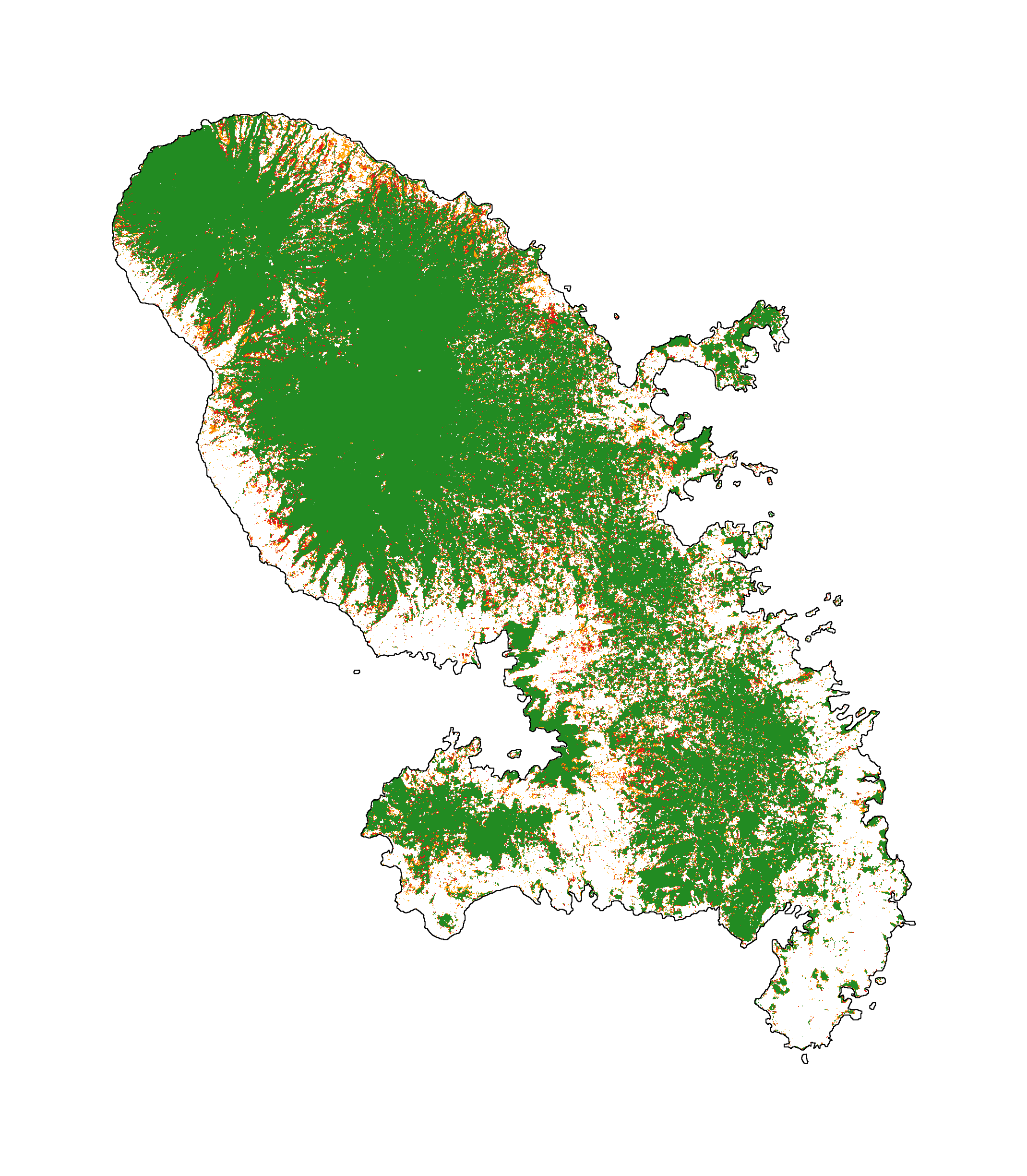
We can plot the past deforestation for the period 2000–2010–2020:
# Plot forest
ofile = "output/nb_ctrydata_fcc123.png"
fig_fcc123 = far.plot.fcc123(
input_fcc_raster="data/forest/fcc123.tif",
maxpixels=1e8,
output_file=ofile,
borders="data/aoi_proj.shp",
linewidth=0.3,
figsize=(6, 5), dpi=500)
ofile
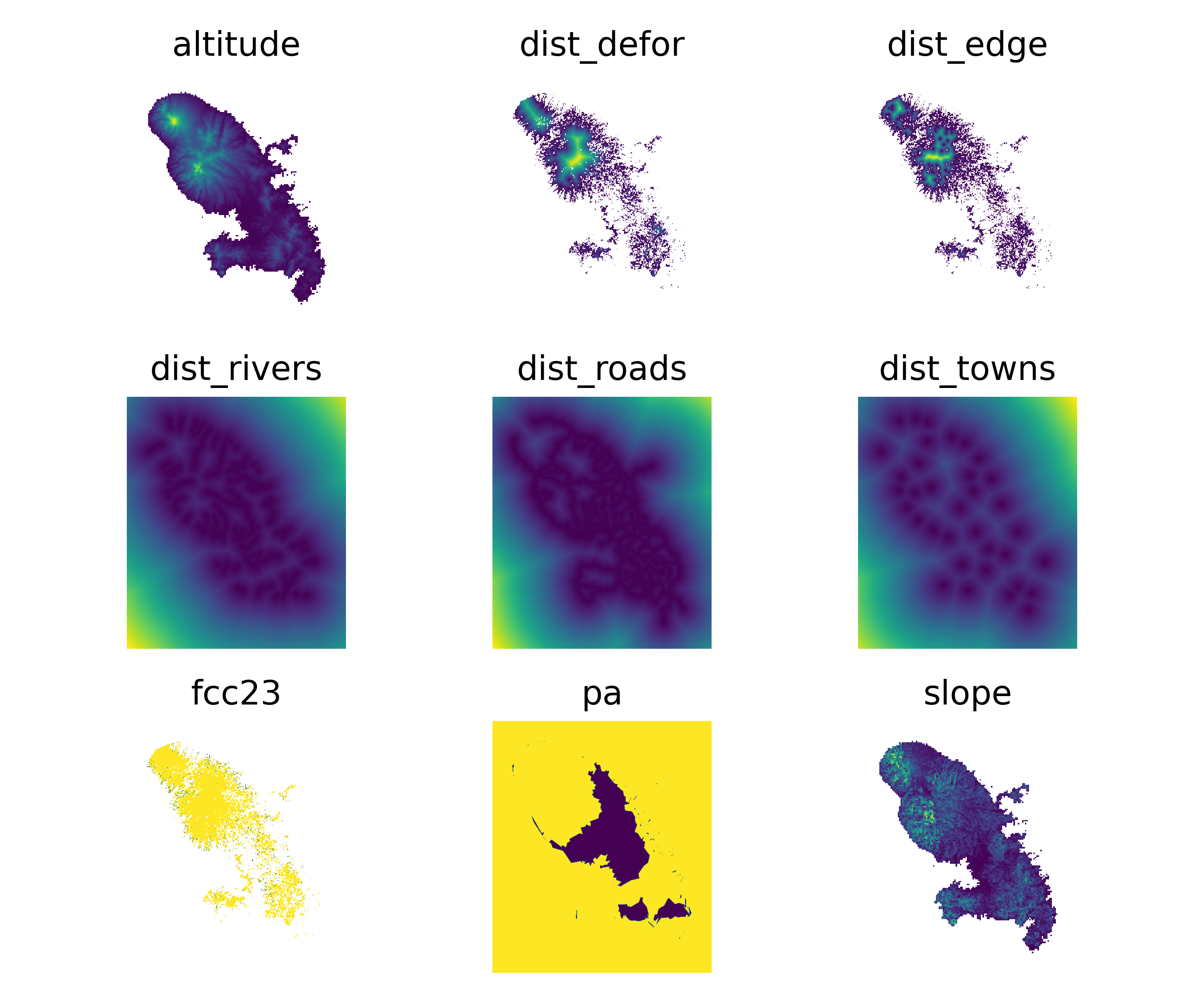
We can also plot the explicative variables:
# Plot explicative variables
ofile_pdf = "output/nb_ctrydata_var.pdf"
ofile = "output/nb_ctrydata_var.png"
fig_var = far.plot.var(
var_dir="data",
output_file=ofile_pdf,
figsize=(6, 5), dpi=500)
fig_var[0].savefig(ofile)
ofile